All published articles of this journal are available on ScienceDirect.
Understanding Personalized Training Responses: Can Genetic Assessment Help?
Abstract
Background:
Traditional exercise prescription is based on the assumption that exercise adaptation is predictable and standardised across individuals. However, evidence has emerged in the past two decades demonstrating that large inter-individual variation exists regarding the magnitude and direction of adaption following exercise.
Objective:
The aim of this paper was to discuss the key factors influencing this personalized response to exercise in a narrative review format.
Findings:
Genetic variation contributes significantly to the personalized training response, with specific polymorphisms associated with differences in exercise adaptation. These polymorphisms exist in a number of pathways controlling exercise adaptation. Environmental factors such as nutrition, psycho-emotional response, individual history and training programme design also modify the inter-individual adaptation following training. Within the emerging field of epigenetics, DNA methylation, histone modifications and non-coding RNA allow environmental and lifestyle factors to impact genetic expression. These epigenetic mechanisms are themselves modified by genetic and non-genetic factors, illustrating the complex interplay between variables in determining the adaptive response. Given that genetic factors are such a fundamental modulator of the inter-individual response to exercise, genetic testing may provide a useful and affordable addition to those looking to maximise exercise adaption, including elite athletes. However, there are ethical issues regarding the use of genetic tests, and further work is needed to provide evidence based guidelines for their use.
Conclusion:
There is considerable inter-individual variation in the adaptive response to exercise. Genetic assessments may provide an additional layer of information allowing personalization of training programmes to an individual’s unique biology.
1. INTRODUCTION
Conventional exercise prescription is comprised of blanket advice. For example, the American College of Sports Medicine (ACSM) recommend >150 minutes of moderate-intensity and >75 minutes of vigorous-intensity cardiovascular exercise per week, along with resistance training twice per week with repetition ranges of 8-12 for novices and 1-12 for intermediates [1, 2]. Within this advice is the implicit assumption that humans respond in a predictable nature to known exercise inputs. Given these recommendations, you might think that an individual’s adaptive response to an exercise intervention is a predictable, standardised phenomenon tightly distributed around an averaged group mean. Yet, in recent decades, studies designed to examine the individual adaptive response to exercise have illustrated large inter-individual variations, in both the magnitude and direction of the resulting response, exceeding far beyond both the expected day-to-day biological perturbations [3. 4], and our conventional perspectives [5, 6].
Historically, training theory has been founded on the implicit, and previously unexamined, assumption that adaptation to training is both standardised and predictable across individuals. Such assumptions form the conceptual bedrock of periodisation theory and exercise prescription literature within sports coaching, public health and medical domains [1, 2, 7-9]. The literature guiding these recommendations typically report group norms which obscure the individual variation that occurs between subjects. However, over the course of the past two decades, evidence demonstrating the unexpectedly extensive inter-individual variation in response to similar training stimuli has accumulated exponentially [5, 6, 10-12]. The illustration of such wide-ranging variations in response conflicts with the traditional exercise prescription assumptions, questioning many of the traditional components of physical preparation.
Resolving the conceptual deficit between current evidence and conventional theory requires an understanding of the influences driving inter-individual response. These influences emanate from diverse academic domains, and the focus of this article is to identify those that interact to customise the inter-individual adaptive response. Furthermore, whilst past reviews have highlighted some of these influences [6], we add findings from the emerging field of epigenetics. Finally, we outline how these influences integrate, and suggest how an enhanced understanding of the broad range of factors influencing adaptive response can help contextualise the limits, and potential value of, emerging gene profiling technologies.
2. INTER-INDIVIDUAL VARIATION IN RESPONSE TO TRAINING
It is well established that, when subjected to the same stimulus, there is a wide variety in response within subjects. Large variations in muscle size (from -2% to +59%), changes in one-repetition maximum (1RM; 0% to +250%) and changes in maximum voluntary contraction (MVC; -32% to +149%) have been reported following 12-weeks of resistance training [10]. The same is true for aerobic capacity improvements, with the HERITAGE (HEalth, RIsk factors, exercise Training And GEnetics) family study finding a mean improvement in VO2max of 384 mL O2 min-1 following 20-weeks’ training. However, some subjects saw no improvement, whilst a small number of subjects saw much larger improvements than the average, as high as 1100 mL O2 min-1 - almost four times the mean [5]. Other studies have reported large variations in response to high-intensity interval training [11], fat loss [13, 14], other health-related aspects including insulin sensitivity, blood pressure, and cholesterol levels [5], and response to ergogenic aids [15].
The individual response to exercise appears to be modality specific. Karavirta et al. [16] randomised 175 subjects into four groups; endurance training only, strength training only, concurrent strength and endurance training, and controls. All groups exhibited a large range in exercise response, with improvements in VO2peak ranging from -10 to +60% in the endurance trained group, and MVC improvements ranging from -15 to +60% in the strength trained group. But it is the strength and endurance trained group where the crucial data lies; although some subjects saw a negative training response in either VO2peak or MVC, no subject had a negative response in both. Additionally, no subject was in the highest quintile of improvement for both VO2peak and MVC. Hautala et al. [17] found that when individuals were given both an endurance and resistance training block, improvements in VO2max differed between modalities. Perhaps most promising was that those seeing the lowest VO2max improvement following endurance training saw a greater VO2max improvement following resistance training. This indicates there aren’t global responders and non-responders to exercise, merely responders and non-responders to specific exercise types. There are thousands of biochemical adaptations to exercise, and a multitude of different training modalities, making it unlikely that there are individuals who see no improvement at all following exercise [18]. This is not necessarily a consensus, however, with others finding that, whilst the inter-correlation in non-response to exercise between exercise modalities is low, it is not zero [19]. Exercise adaptation occurs through several separate pathways specific to each training modality. The lack of global non-responders suggests the driver of individual variation in exercise response could be down to variation within these pathways. This variation would likely occur due to genetic and non-genetic influences, which combine to create a unique adaptive outcome.
Outside of specific performance-enhancing adaptations following exercise, there is also a large range in terms of health improvements. Returning to HERITAGE, the mean improvement in insulin sensitivity within a sub-cohort was 10% [5]. However, this varied between subjects from large improvements, to no change, to a decrease in insulin sensitivity. Factors affecting this individual response included gender, race, and starting body weight [20]. Similarly, disease state can also modify improvements in insulin sensitivity following exercise training, with type-II diabetics and non-diabetics seeing improvements of differing magnitudes [21].
Even very complex traits, comprised of many factors, show large ranges in variability between subjects. Injury risk in sport is multifactorial, comprised of internal and external risk factors, along with an inciting event [22]. Even within this complex model, not all high-risk athletes will get injured, nor will low-risk athletes avoid injury [23]. The same is true when looking at response to a dietary supplement, such as vitamin D, with genotype, baseline serum 25-hydroxyvitain D (25(OH)D) levels and body mass index (BMI) all modifying increases in serum 25(OH)D levels following supplementation [24]. A large individual variation in the ergogenic effect of caffeine has also been widely reported [15, 25, 26].
3. POTENTIAL MECHANISMS DRIVING THE INDIVIDUAL RESPONSE
Given that these large variations in exercise adaptation exist between subjects, it is necessary to identify and understand the causes of this variation. Here, we outline how proposed mechanisms interact to lead to the observed extensive inter-individual variation in exercise adaptation.
3.1. Genetics
Following the completion of the Human Genome Project in 2003, genetic analysis has become increasingly affordable, making research into the effects of genes on fitness and performance more feasible. Knowledge of these genetic influences has progressed significantly in recent years, moving from the idea that all traits are determined by a single gene (which holds true in select disease states such as Cystic Fibrosis [27] and Huntington’s disease [28]), to more complex polygenic interactions. The “single gene as a magic bullet” philosophy has previously been present in sport [29], with some coaches believing that individual genes are responsible for athletic performance. However, no single gene has been discovered. Instead, we are faced with the reality that elite athletes possess many favourable alleles [30, 31] with no athlete possessing the perfect genetic profile for elite performance [32].
All traits, therefore, exist on a spectrum; from single gene traits at one end to complex polygenic traits at the other. Whilst it might be thought that complex traits can never be fully understood regarding their genetic component, research has identified candidate genes associated with highly complex traits such as intelligence [33], educational attainment [34], height [35], and chances of being an elite athlete [36]. Of course, these traits are also dependent on non-genetic factors, but there is an inherent genetic component within them. Returning to exercise adaptation, the heritable component differs from trait to trait. For example, the results of HERITAGE indicate approximately 50% of heterogeneity in VO2max improvement following training is determined by heritable factors [37], whilst muscle fibre type is 45% to 99.5% heritable [38, 39], and 52% of muscle strength phenotype is heritable [40]. Knowledge of genes affecting this response may allow for manipulation of training factors such as volume, intensity, frequency and rest-periods to improve exercise response. Indeed, recent research has argued whether true non-responders to specific exercise modalities exist, with increases in exercise intensity and frequency eliminating exercise non-response [41-43].
3.1.1. Gene Polymorphisms & Exercise Adaptation
At least 120 genetic markers are linked to elite athlete status [44], with approximately 10% of these replicated in at least three studies; yet more genes are implicated with exercise adaptation [45]. Elite athletes are a good start point in the search for candidate genes driving exercise response, as they represent a highly specialised cohort. For example, elite sprinters are likely very good at sprinting because they possess genotypes predisposing favourable adaptations following speed-power training. One such gene is ACTN3, which encodes for alpha-actinin-3, a protein that forms part of the Z-line in muscle fibres. A single nucleotide polymorphism (SNP) within ACTN3, known as R577X, arises from a C → T substitution, resulting in a premature stop codon (X) in place of arginine (R). Approximately 18% of individuals are homozygous for the X allele, causing a deficiency of alpha-actinin-3 [46]. Whilst this isn’t associated with any disease state, it does mean that XX genotypes tend to have a lower percentage of type-IIx muscle fibres [47]. The X allele is uncommon in elite speed-power athletes, and potentially more common in elite endurance athletes when compared to controls [48]. This indicates the XX genotype is unfavourable for elite power performance, but potentially has a beneficial effect on endurance performance. Subsequent research has confirmed the association between the R allele and power performance, although the link between the X allele and endurance is less clear [49]. Other replicated SNPs found to affect athletic performance include ACE I/D [50-52], PPARGC1A Gly482Ser [53-55], GABPB1 (rs7181866) [56, 57], BDKRB2 +9/-9 [58, 59], and HIF1A Pro582Ser [60, 61].
After identifying a relevant polymorphism, the next step is to elucidate how this polymorphism affects individual training response. With ACTN3, RR genotypes show greater improvements in peak power and strength than XX genotypes following resistance training in elderly populations [62, 63]. The mechanisms driving these differences are not fully understood. Increases in mTOR and p70S6k, stimulators of skeletal muscle hypertrophy, have been found to be greater in R allele carriers than XX genotypes following high-intensity exercise [64], and testosterone levels may be higher in RR genotypes [65]. R allele carriers also tend to have a higher percentage of type-II muscle fibres [66], potentially allowing greater hypertrophy following resistance training [67, 68]. These findings may go some way to explaining the differences in training responsiveness between ACTN3 genotypes. Again, similar research has shown a modifying effect of other polymorphisms on training response, including ACE I/D [69-71] and PPARGC1A Gly482Ser [72-74], as well as the influence of genetic variation on other traits, including injury risk [75-77] and exercise recovery [78, 79].
3.2. Environmental Factors
If heritable factors are responsible for a part of exercise adaptation, the obvious question to ask is - what is responsible for the other part? These non-genetic factors are often termed “environmental”, which we will define as non-genetic factors. Within this review we will divide them into four groups; individual history, programme design, psycho-emotional factors, and nutrition. These non-genetic factors can be both acute, affecting a single or small number of consecutive sessions, or chronic, affecting response to the training programme as a whole.
3.2.1. Individual History
A phenotype is the observable expression of an individual’s genotype, which is impacted by that person’s environment [80]. Within this paper, we can consider individuals to have either a highly-, normal-, or under-adaptive phenotype, influenced by their genotype (see Gene polymorphisms and exercise adaptation), but also environmental variables. One such variable is baseline fitness, which impacts recovery from exercise [81-83]. Another is previous training history, with trained individuals showing differences in adaptive mechanisms post-exercise compared to beginners [84], and subject age [85]. When looking at dietary interventions, diet history can modify responsiveness to interventions, with previous weight loss attempts potentially making future weight loss harder [86]. Finally, higher habitual physical activity can enhance the response to endurance training [87]. Within HERITAGE, correlates of VO2max improvements following training included baseline VO2max, age, gender, weight, ethnicity, and achievement of target workload [88]. Baseline phenotypes appear to influence separate traits to differently, comprising a smaller portion of VO2max improvements following exercise (11-16%) and a larger portion of blood pressure response following exercise (21-47%) [6].
3.2.2. Programme Design
Training programme design (exercise selection, frequency, duration, intensity, recovery times, repetition and set ranges, etc.) can also influence the magnitude of adaptation to training [67, 68, 89-93], as can time of day [94.95], such that two people with an identical genotype doing different training programmes would see a difference in phenotype. Indeed, increasing total exercise volume, frequency and intensity reduces, and perhaps eliminates, exercise non-response, suggesting that environmental influences can perhaps over-ride the genetic pre-disposition to exercise non-response [41-43, 96].
3.2.3. Psycho-Emotional Factors
In recent years, attention has turned to how the brain influences exercise performance and adaptation. Initially, this focused on fatigue, with both the Central Governor [97] and psychobiological [98] models proposed to explain the relationship between brain and fatigue. Following this, interest has built around understanding the relationship between brain and physiology, especially regarding exercise adaptation. Previous work has indicated that response to a stressor - including exercise, feelings of fatigue, and pain – is filtered through the brain’s emotional centres, which evaluate the stressor in terms of its threat [99]. Current perceptions suggest that biological adaptation, to an imposed or perceived stressor, is not regulated by the magnitude of that stressor, but by the nature of the stress response launched to remediate the challenge presented to the neuro-biological system [101]. The nature and magnitude of the stress response is hence governed primarily by the emotional resonance afforded the perceived threat presented by the stress-inducing event. This emotional interpretation subsequently initiates a neuro-chemical response, proportional in magnitude to the perceived threat presented by the stressor. This neurochemical response, in turn, launches the cascade of downstream bio-chemical responses which subsequently drive all peripheral adaptations [99-102]. From this, we can see that the magnitude of response to a stressor is not solely dependent on the stressor itself, but the emotional resonance attached to this stressor; this emotional response drives the bio-chemical and hormonal alterations, in turn driving all subsequent physiological and peripheral tissue adaptations.
This emotional response is complex, and is best summarised by Ganzel et al. [100] In their model, the authors describe the factors that mediate the emotional response, including prior context, such as previous traumatic experiences, evolved coping behaviours, and health. This prior context interacts with the current state of the organism, both in terms of emotional state (influencing prior mental health, which influences acute emotional response to a stressor) and, via the chemical changes that drive subsequent physical responses, physical changes that accompany chronic stress, such as increased cortisol (which influences prior physical health, itself a modifier to the acute response to a stressor). These factors combine to influence the weighting the emotional system places on an acute stressor, affecting the acute physical response.
By combining the work in this field, we can summarise that every stressor, including exercise, exerts a neurological, biological, psychological and emotional load depending on individual interpretation [99, 100, 103]. This means that what often feels solely like a physical response, such as fatigue, is mediated by perception, suggesting the psychological and biological responses to a stressor are irrevocably mutually entwined.
3.2.3.1. Factors Affecting Psycho-Emotional Response
The response to a stressor is altered by both environmental and genetic factors. These include lack of sleep, which impacts exercise recovery [104, 105], promoting the release of stress hormones [106]. This potentially leads to a loss of aerobic [107] and strength [108] ability, and increases the inflammatory response [109], altering training performance and hence adaptation. Sleep restriction, both acute and chronic, can alter the perception of a stressor [110], modifying the psycho-emotional response [111].
Stress interpretation is modified by heritable factors, including polymorphisms in genes such as COMT [112], BDNF [113, 114], and 5HTTLPR [113] that impact the stress response, altering exercise adaptation and performance [115, 116]. The microbiome, which is influenced by both environmental and genetic factors, also affects the stress response in athletes [117]. Finally, epigenetic modifications (see Epigenetics) impact stress interpretation pathways [118], explaining how childhood trauma influences adult stress behaviours [119].
The individual stress response also impacts the adaptive mechanisms following exercise. Psycho-emotional stress influences exercise adaptation by decreasing immunity and recovery [120], and increasing the risk of injury [121]. In addition, baseline stress has been correlated with VO2max improvements [122]. Given that the stress response is partially hormone led [123], and these hormonal changes can be fast-acting [124], the stress state of the subject at the time of exercise can modify the adaptive response both acutely and chronically [125]. As an example, subjects with lower stress scores show greater increases in both bench press and squat strength compared to subjects with higher stress scores [126]. Similarly, an athlete who has just argued with a spouse and has long-term financial worries is less likely to mount an optimal adaptive response than a content athlete [99].
The acute psycho-emotional response to exercise can cause variation in work-rate within that session, contributing to the inter-individual variation in exercise response [88]. Within-session work-rate is comprised of various factors, including residual fatigue, but also via psychological factors that impact within-session work via the psychobiological model [98]. Individual variation in perception of work-rate can lead to changes in exercise performance [127], and this perception of work rate is influenced by a myriad of factors [97]. Perception of effort also has a heritable component, which explains 35% of the variance in rating of perceived exertion (RPE) between subjects [128].
Finally, the placebo effect, expected outcomes, and previously-held beliefs alter the emotional evaluation of a stressor, modifying training performance and adaptation. An ever-increasing body of literature illustrates that a subject’s prior beliefs alter performance, including belief that they have taken caffeine [129], sodium bicarbonate [130], and doping agents [131-133]. Returning briefly to sleep, “placebo sleep” can improve cognitive function [134], again illustrating the power of belief. Given that expected beliefs can alter effort within a training session, whether a subject believes exercise is positive can affect the outcome of exercise- and nutritional-intervention trials [135, 136]. Emerging research seems to suggest that certain genotypes are more sensitive to expectancy, placebo and nocebo effects [137], again illustrating the consistent underlying influence of genetics on environmental factors.
3.2.4. Nutrition
An additional factor that influences exercise adaptation is nutrition. Macronutrient intake impacts both exercise performance and adaptation [138-140]. The same is true for micronutrients; for example, serum vitamin D levels are associated with muscle power and force, both acutely [141] and in response to a training programme [142]. Recently, attention has focused on individual variation within the gut microbiota, which impacts post-exercise recovery and mood states, altering adaptation [117, 143]. Finally, long-term high dose antioxidant use may blunt the adaptive response to exercise [144, 145], leading to the possibility that differences in dietary composition could contribute to the inter-individual variation in exercise response. Other nutritional factors modifying the acute physiological stress expected following training include short-term macronutrient intake [146], antioxidant intake [147, 148], and use of medications such as non-steroidal anti-inflammatory drugs (NSAIDS) [149-151].
These nutritional factors are influenced by genetic variation. The microbiota, for example, is influenced by host genetics [152]. Returning to the vitamin D example, polymorphisms in genes, including VDR, influence muscle strength [153], which in turn influences training response. VDR can also alter vitamin D requirements [154]. Vitamin D supplementation may also enhances improvements to a strength training programme [142], which begs the question - do non-responders to strength training not respond because of genetic factors, or is their response blunted due to vitamin D insufficiency (which in turn can be influenced by SNPs)? Given that nutrition impacts gene signaling post-exercise [155, 156], it’s easy to see how both genes and environment combine and interact to create the phenotype.
Finally, the use of ergogenic aids alters the performance level within individual training sessions, in turn affecting the long-term adaptation that accumulates over time. One such aid is caffeine, which has a clear, replicated ergogenic effect on exercise performance [157-159], the effects of which are modified by genetic variation [26, 160]. Another is creatine, which can affect intra-session recovery, allowing for a greater workload to be completed [161].
3.3. Summarising Gene-Environment Interactions
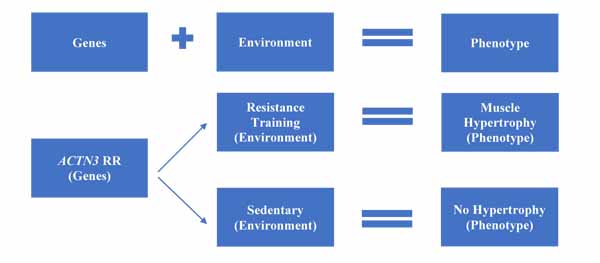
Having discussed the different genetic and environmental aspects that affect exercise response, it is worthwhile summarising these within a model. Fig. (1) shows the typical gene-environment model, where both genetic and environmental factors interact in an additive manner to determine the post-adaptation phenotype. As a simplified example, two individuals who are homozygous for the R allele of ACTN3 will have different phenotypes based on environmental factors. If subject A undertakes high-load resistance training, they will likely see good levels of muscle hypertrophy. If subject B is sedentary, then they won’t see hypertrophy, no matter how favourable their personal genotype.
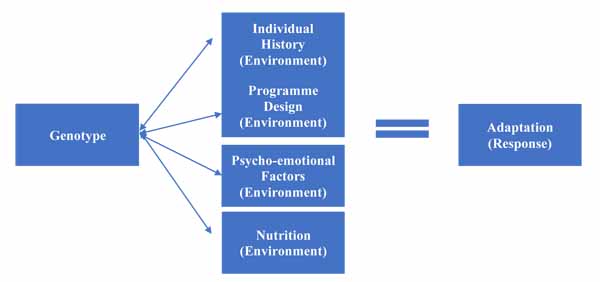
However, as explored in environmental factors, there are a variety of environmental factors that affect training response. These have a complex relationship with genetic factors; they can affect genetic expression, but are also affected themselves by SNPs within specific genes. This allows us to create a more complex model, as per Fig. (2), which illustrates the increasing complexity.
3.4. Epigenetics
Having introduced genetics and environment, two aspects that we typically think combine to create the phenotype, we turn our attention to epigenetics. Epigenetics refers to changes in gene function that occur without a change in the nucleotide sequence [162]. These changes can be heritable, but also changeable over the course of time within an individual [163], influenced by both genetic and non-genetic factors. The three main epigenetic mechanisms are DNA methylation, histone modifications, and non-coding mRNA, and all act as a way for our environment, through factors discussed previously, to impact genetic expression.
3.4.1. DNA Methylation
The most extensively studied epigenetic mechanism, DNA methylation occurs through the addition of a methyl group to a cytosine base [164], making that section less accessible for translation [165]. This can be positive or negative depending on whether expression of that gene is desired; methylation of oncogenes and obesity-risk genes is likely positive, whilst methylation of tumor suppressor genes, and those driving exercise adaption is less ideal [166, 167]. The same stimulus can both increase and decrease in methylation in different genes [168]. For example, six-months of aerobic exercise lead to decreases in methylation (hypomethylation) of muscle genes, promoting adaptation [167], and increases in methylation (hypermethylation) in adipose tissue genes, potentially stimulating weight loss [169]. Similarly, PPARGC1A, a gene influencing mitochondrial biogenesis [170], exhibited increased methylation following nine-days of bed rest, and decreased methylation after four-weeks of re-training [171].
DNA methylation is modifiable within an individual; the methylation profiles of obese patients become more like lean subjects’ following a weight-loss intervention, for example [172]. The levels of methylation in response to the same stimulus may also change over time, with higher levels seen in elderly subjects’ post-exercise, possibly due to accumulation of aberrant methylation in these subjects that needs correcting with exercise [173]. Of the three epigenetic modifications detailed here, methylation is the most stable [164], with early life experiences – even those pre-birth – having a long-term effect on gene expression [174]. Methylation patterns can even be passed down generations, raising the possibility that methylation markers affecting elite athlete status and fitness may be partially inherited [168].
3.4.2. Histone Modification
Our DNA is coiled around histone proteins, giving it a specific shape. The epigenetic variation caused by histone modification occur via acetylation of this structural histone protein, changing its shape. This makes the DNA comparatively easier to read, increasing the expression of these genes [175]. Histone acetylation is controlled by histone acetyl-transferases (HAT), whilst histone deacetylases (HDAC) removes the acetyl group, reducing translation at that point [176]. In mice, the presence of a specific HDAC (HDAC5) can reduce the adaptations expected following exercise [177], illustrating how histone modifications might affect exercise response. In humans, HDAC5 levels are lower following training, confirming that these proteins play a role in exercise adaptation, although at present the causes of individual differences in HDAC5 levels are not clear [175]. Histone modifications are constantly in a state of flux, making them the most transient of the epigenetic changes [164].
3.4.3. Non-Coding RNA
RNA is typically used by the body as messenger RNA (mRNA), passing information from DNA to the ribosomes, where protein synthesis occurs. However, most RNA within the body is non-coding; instead, it regulates genetic expression or catalyses chemical reactions [178, 179, 180]. Within epigenetics, of interest is micro RNA (miRNA), molecules which exert control over mRNA, either by inhibiting translation or causing degradation before translation occurs [181]. This indicates miRNA could regulate gene transcription post-exercise, affecting adaptation. In subjects matched for diet, training history, age and body mass, a 12-week resistance training programme elicited adaptations of differing magnitude, partially mediated by specific miRNAs; levels of these miRNA were correlated with greater adaptations, including increases in strength [180]. miRNA has also been reported to influence aerobic exercise adaptation [180-183]. It’s not clear at present what factors affect circulating levels of miRNA, making it difficult to harness this knowledge at present.
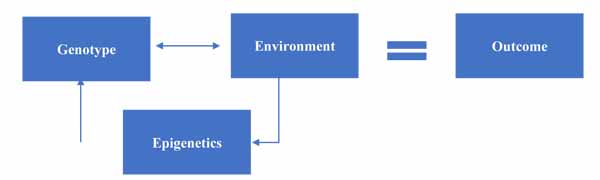
At this point, we can update our model to include the impact of epigenetics on gene-environment interactions, as seen in Fig. (3).
3.4.4. Genetic Influences on Epigenetic Modifications
So far, we’ve covered how epigenetic mechanisms allow for the environment to impact genetic expression, which is typically how epigenetics is viewed. However, genetic variation can also affect the efficiency of epigenetic modifications, bringing things full circle. This is most well established in terms of methylation, where several genes [184], including MTHFR [185], affect DNA methylation, in turn affecting epigenetic modifications post-exercise. Elite athletes have a greater number of polymorphisms across several genes that affect methylation status, resulting in a genetic predisposition to hypomethylate, and this lack of methylation potentially increases post-exercise muscle hypertrophy by increasing specific gene transcription [186, 187].
3.4.5. Environmental Influences on Epigenetic Modifications
Along with genetics, environmental influences such as nutrition can alter epigenetic modifications. For example, a high calorie diet appears to increase methylation of genes controlling metabolism, making metabolic dysfunction more likely [188]. As discussed, genes influence the efficiency of methylation, but also interact with environmental factors to control these changes, adding an extra layer of complexity. As an illustration, MTHFR encodes for an enzyme that coverts the folate derivate 5,10-methylenetetrahydrofolate (5,10MTHF) to 5-methylterahydrofolate (5MTHF), creating s-adenosylmethionine (SAM) – the agent for DNA methylation. Simply put, this pathway starts with folate, which is converted to intermediaries by MTHFR, with the availability of these intermediaries affecting methylation efficiency [189, 190]. The two common MTHFR SNPs, C677T and A1298C, influence the activity of the MTHFR enzyme. Focusing on C677T, T allele carriers typically have poorer conversion of folate, which influences methylation. When placed on a low folate diet (≈115μg/d) for seven weeks, subjects show a decrease in methylation. This effect is greater in TT genotypes, and was reversed after seven-week high folate diet (≈400μg/d) [190].
Exercise is another environmental influence that impacts epigenetic changes through alterations in gene silencing and expression [191]. The homeostatic stress caused by exercise drives epigenetic modifications [192], which in turn can lead to exercise adaptations by increasing translation and transcription of proteins involved in adaptive mechanisms, including AMPK and PGC-1a [193].
Environmental influences on epigenetic modification can play a big role in determining an individual’s phenotype; in individuals with the same genotype (monozygotic twins), differences in environment lead to different epigenetic changes [194], altering type-II diabetes risk, for example [195]. Alongside nutrition and exercise, other environmental factors that influence epigenetic modifications include psychological trauma [196, 197], which can be passed down generations [198], but also reversed [199]. Environmental toxins such as tobacco smoke, dietary polyphenols, alcohol and shift work all also impact epigenetic regulation [200].
Having reviewed the impact of both genetics and environmental factors on epigenetic modifications, we can add these factors into a final model, discussed in section 4.
4. A FINAL MODEL TO EXPLAIN THE CAUSES OF INTER-INDIVIDUAL VARIATION
As detailed earlier, genetics can clearly impact the magnitude of adaptation to exercise, both in single SNP/gene (e.g. ACTN3) and gene-combination (e.g. HERITAGE) models. Previously, we examined non-genetic aspects influencing this response, including nutritional status and training history. As an example, vitamin D status can impact performance gains [142], and vitamin D status itself is modified by sunlight exposure and supplementation [142], but also gene polymorphisms [24, 154]. The same is true for a number of other nutrients; polymorphisms within HFE [201], a gene that impacts iron status alongside dietary iron intake [202, 203], may also impact improvements in aerobic fitness following training [204]. It is clear, therefore, that genetic and non-genetic factors are linked. The same is true for acute environmental factors, such as a stressor, which we suggested might affect response to a single exercise bout. These acute factors are also influenced by genetic factors, such as a SNP in COMT, that modulates stress response [205]. They are also influenced by environmental factors such as previous trauma [206].
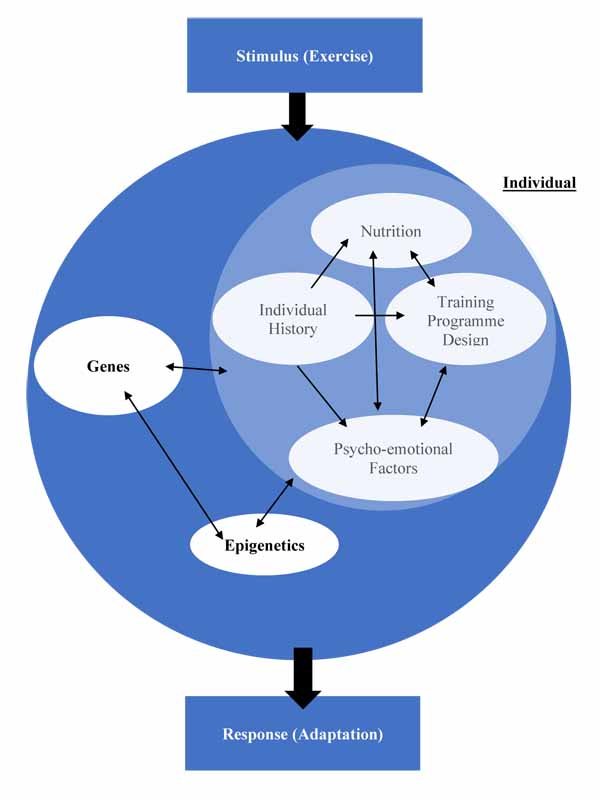
Having then introduced epigenetics, the mechanism through which environmental aspects influence genetic expression; we explored how genetic and non-genetic factors also influenced epigenetic modifications, further illustrating the complex relationship between all factors, requiring an update to the model proposed in Fig. (3). This culminates in a model illustrating how these factors interact, creating a unique outcome for each individual in response to a stimulus. This response is not stable, as the component factors themselves can be highly variable over time; just because an individual saw a performance improvement after one training programme doesn’t guarantee the same improvement following the same programme once more [207]. Fig. (4) illustrates this complex relationship.
5. HARNESSING THIS KNOWLEDGE TO IMPROVE PERFORMANCE
Having discussed the main aspects that affect individual adaptation to exercise, it’s crucial to make this information usable to athletes. Competing at the highest level is a function of talent alongside optimal training – but how does an athlete know their training is optimal? Typically, this requires trial and error, which is costly in terms of both time, and, if the trial is ineffective, performance. Given that athletes only have a window of a few years to compete at their peak, time spent undertaking sub-optimal training can be damaging. Having more information on which to base decisions regarding training methodology would be very attractive to everyone involved in sport. Currently, most tests carried out on athletes are phenotypic, such as VO2max and vitamin D tests. This testing has use, providing a snapshot of where the athlete is at a point in time and informing training requirements, but has minimal long-term predictive ability.
Given that a large proportion of inter-individual variation is down to genetic factors, testing for these factors holds promise. This could be single gene/SNP testing, or, more promisingly, larger scale testing such as whole genome sequencing. The cost of these tests has dropped in recent years, increasing accessibility [208]. This gives rise to the potential use of genetic tests to inform training programme design, which may have predictive ability [183, 209, 210]. Whilst single gene models might give some insight into exercise response [62, 211], adaptation to exercise is not determined by a single gene. Instead, groups of genes influence the various cellular pathways controlling adaptation [212]. By examining just one gene, such as ACTN3, we run the risk of ignoring the effects of these other genes. One way to overcome this is to use a multi-gene model, comprised of an algorithmic approach that allows for the evaluation of many gene polymorphisms.
One method used in this regard is the Total Genotype Score (TGS). This method has been used against retrospective data to improve identification of at-risk individuals for cardiovascular disease and type-II diabetes [213, 214]. Within the sports world, it has thus far been examined primarily as a potential tool for the discovery of elite athletes [30, 31, 215] although the consensus is that there is currently no predictive ability of genetics in the identification of elite athletes [216]. Pooled data from three independent aerobic training programmes – HERITAGE, DREW and STRRIDE – showed that those with a TGS of ≥19 had VO2max improvements 2.7 times greater than those with a score of ≤9, although this was conducted post-hoc and not used to inform programme design [217]. Presently, the use of a TGS or other algorithm has not been widely utilised in regards to interventions to improve exercise response. One study used a TGS to retrospectively explain training response over the career of an athlete [218]. Another used a weighted algorithm to personalise an eight-week resistance training programme, with those doing genetically matched training demonstrating significantly greater improvements across tests of power and endurance than those doing genetically mismatched training. In addition, over 80% of subjects identified as high responders were from the matched group, whilst 82% of non-responders were from the mismatched group [209]. This suggests genetic testing might reduce non-response to exercise; something that will excite elite athletes, but which may also have public health connotations in the fight against obesity. Another method combined the use of RNA profiling with SNPs to create a molecular predictor of VO2max response to aerobic training [183], although this has yet to be satisfactory replicated [88]. It is still early in this journey, with a far greater body of research required; nevertheless, it does appear that we are getting closer to being able to utilise this knowledge. Doing so will also require manipulation of training variables, such as exercise intensity, duration, volume, as well as nutritional interventions. It must be remembered that the use of genetic information can better inform these manipulations, but does not replace them.
Genetic testing is somewhat controversial [216, 219, 220], with the controversy comprised of various factors. One of these is the use of genetic testing for talent; certainly, there is no evidence that genetic testing can or should be used in this way [216]. The second is whether they have utility in terms of exercise modification. A recent consensus statement suggests they don’t [216], although no evidence is given within the statement to support this standpoint. It’s certainly true that, at present, only a small number of studies have looked at training modifications based on genetic information, but this number is expected to grow in the future, leading to the possibility that genetic information might have some use alongside other more traditional information sources [221, 222]. Finally, there are ethical aspects to consider and overcome. Should there be a minimum age for genetic tests? Can the results of a genetic test be placed in the correct context for an athlete? Who owns the genetic data, the athlete or the team? If an athlete refuses a genetic test, will they be discriminated against? What happens if a genetic test unearths a potential medical issue, such as increased Alzheimer’s disease risk? These questions, and others, will need to be answered before genetic testing can become widespread in sport. Finally, there needs to be assurances that genetic test results won’t be used for selection purposes, or any other discriminatory practices. If these ethical hurdles can be overcome, there is the potential to use genetic testing in exercise prescription and modification alongside other more traditional aspects.
CONCLUSIONS AND FUTURE DIRECTIONS
Throughout the course of this review, we have explored some of the factors that modify the individual response to a stimulus, primarily exercise adaptation. We’ve seen how our environment can impact adaptation, through aspects such as sleep and nutrition, and we’ve also examined how epigenetic modifications allow communication between the environment and genetic expression. However, a constant theme throughout has been the influence of genetics on the response to a stimulus. Differences in genotype are responsible for a large amount of variation in exercise adaptation, but genetic factors also influence environmental aspects such as nutrition and epigenetic efficiency. Given that genetic factors are such a consistent and fundamental modulator of how someone responds to exercise, knowledge these factors within an individual would prove useful. For the first time, this knowledge is affordable and available through genetic testing, allowing athletes and coaches to have an idea of how they will respond, and to modify training to account for this. The information gained from a genetic test represents an additional piece of information to inform needs much like a vitamin D screen, heart rate variability for recovery, or a 1RM strength test. It is still early in the use of genetic testing for sports people, and a significant body of research is required to identify yet more SNPs involved in exercise adaptation, along with other areas of interest to athletes; injury risk, recovery speed, and the ergogenic effects of nutritional aids. However, research is starting to indicate the utility of these tests. Indeed, some sports teams have been using genetic information [223], but without any evidence-based practice. Given the apparent desire of high level sports people to utilize genetic information to inform programme design, the development of evidence-based guidelines is paramount, which of course means that further research on the potential use of genetic information in training response in required, particularly from a predictive standpoint. As such, further research should focus on:
- Replication of existing, and discovery of further SNPs that impact exercise adaptation.
- Examining the interplay between genes, environment, and epigenetic modifications on exercise adaptation.
- The development of evidence-based guidelines on the use of genetic assessments in sport, with particular reference to ethical considerations.
The ability to harness this information potentially represents a new dawn in understanding exercise adaptation, allowing athletes to better their quest to become faster, higher, and stronger.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
Neither Craig Pickering nor John Kiely have any conflicts of interest pertaining from the publication of this manuscript.
ACKNOWLEDGEMENTS
Craig Pickering is an employee of DNAFit Ltd. He received no financial incentive for the production of this manuscript.


